Publications
You can find a full list of my publications on my Google Scholar profile.
* = equal contribution.
Bayesian optimization
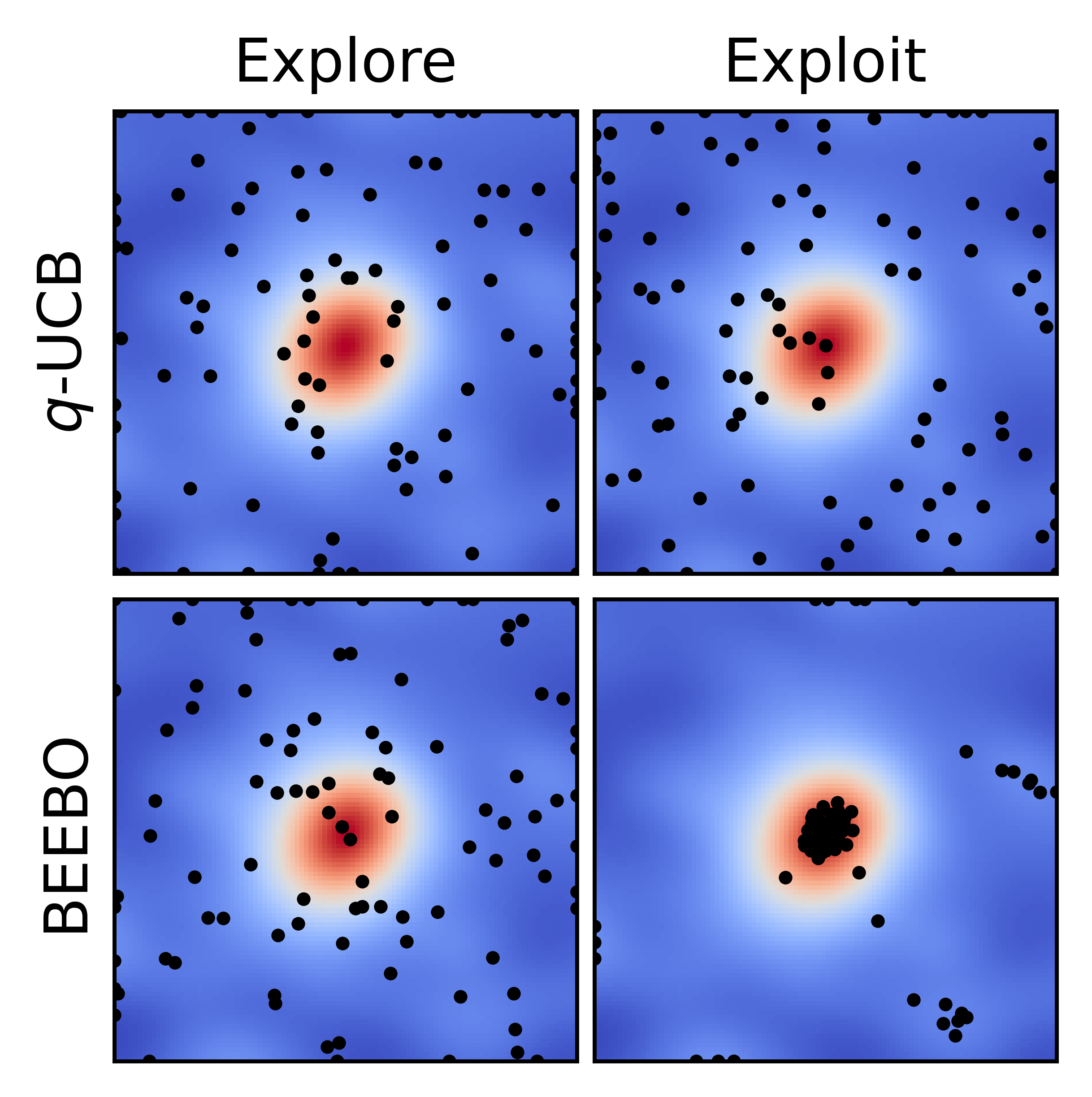 Batched Energy-Entropy acquisition for Bayesian Optimization
Batched Energy-Entropy acquisition for Bayesian Optimization
NeurIPS 2024
Felix Teufel, Carsten Stahlhut, Jesper Ferkinghoff-Borg
A statistical physics inspired acquisition function for Bayesian optimization with Gaussian processes that natively scales to batch acquisition mode. Enables UCB-like control of the exploration-exploitation trade-off at larger batch sizes.
Code on Github
Python package
Representation learning on DNA
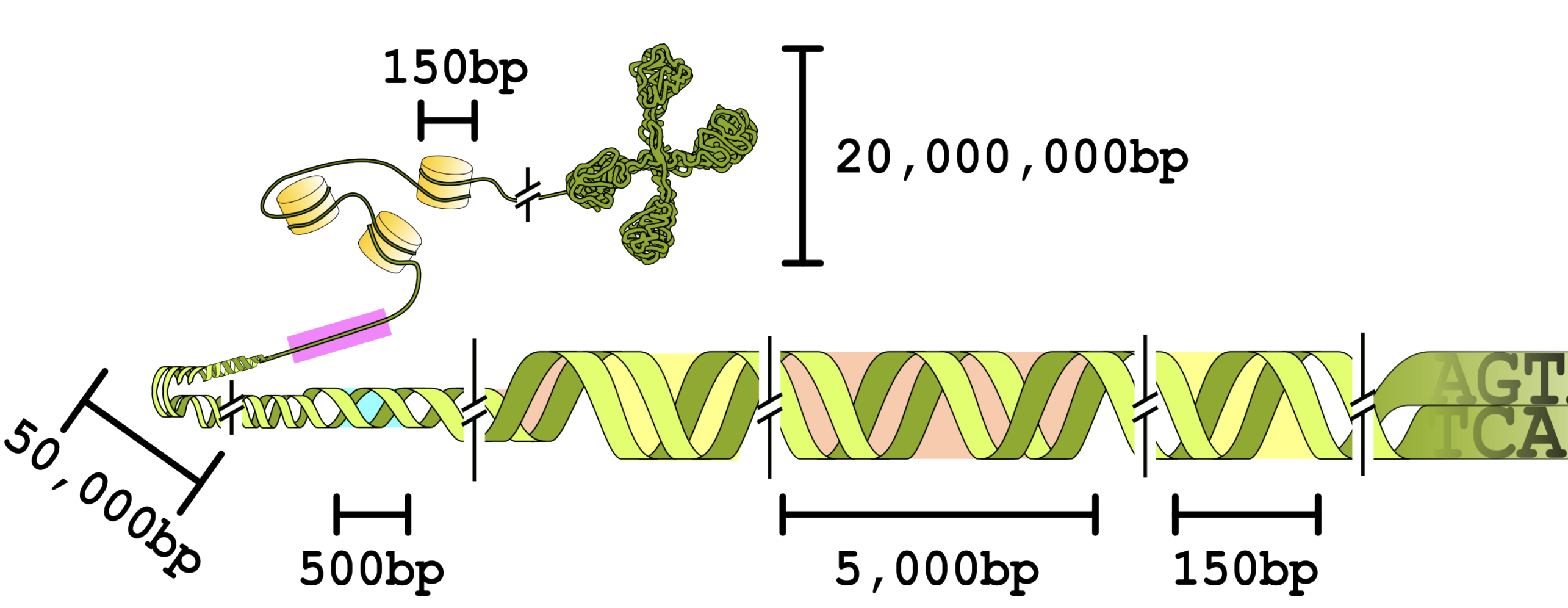 BEND: Benchmarking DNA Language Models on biologically meaningful tasks
BEND: Benchmarking DNA Language Models on biologically meaningful tasks
ICLR, 2024
Frederikke Isa Marin*, Felix Teufel*, Marc Horlacher, Dennis Madsen, Dennis Pultz, Ole Winther, Wouter Boomsma
We introduce BEND, a collection of prediction tasks for evaluating the performance of DNA LM representations over a range of length scales.
Code on Github
Data
Blog post
Protein sorting and location prediction
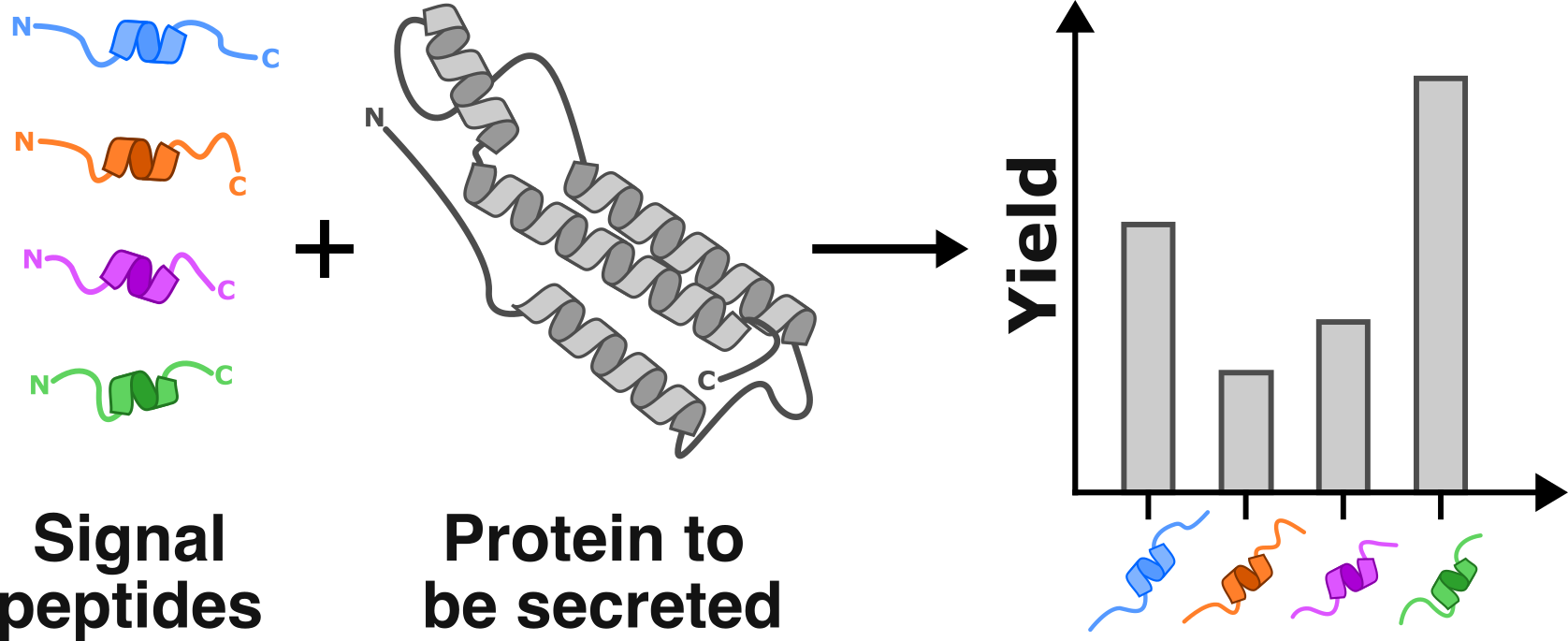 SecretoGen: towards prediction of signal peptides for efficient protein secretion
SecretoGen: towards prediction of signal peptides for efficient protein secretion
GenBio Workshop at NeurIPS, 2023
Felix Teufel, Carsten Stahlhut, Jan Refsgaard, Henrik Nielsen, Ole Winther, Dennis Madsen
SecretoGen is a generative transformer that designs host- and protein-optimized signal peptides. We show that it can be used for ranking signal peptide sequences that have good secretion performance.
Code on Github
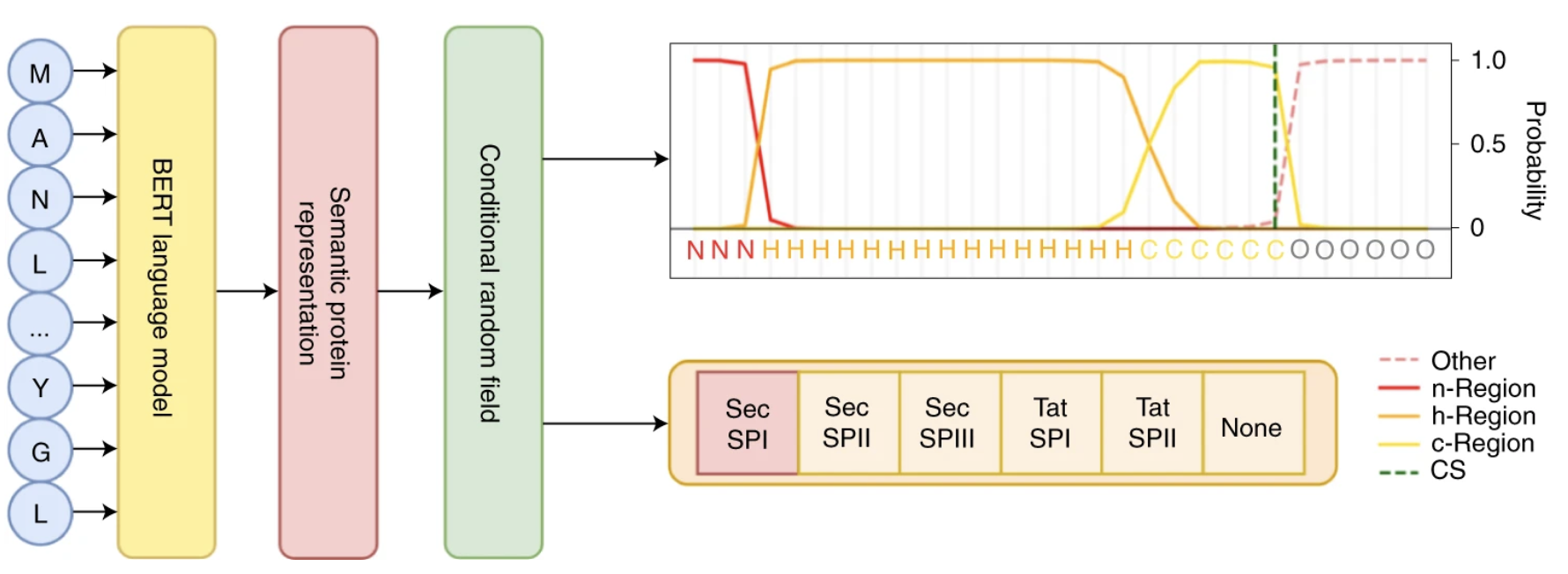 SignalP 6.0 predicts all five types of signal peptides using protein language models
SignalP 6.0 predicts all five types of signal peptides using protein language models
Nature Biotechnology, 2022
Felix Teufel, Jose Juan Almagro Armenteros, Alexander Rosenberg Johansen, Magnús Halldór Gíslason, Silas Irby Pihl, Konstantinos D. Tsirigos, Ole Winther, Søren Brunak, Gunnar von Heijne and Henrik Nielsen
We used protein language models to build SignalP 6.0, the first signal peptide predictor capable of predicting all known types of signal peptides in protein sequences.
Available online at DTU Health Tech
Code on Github
Blog post
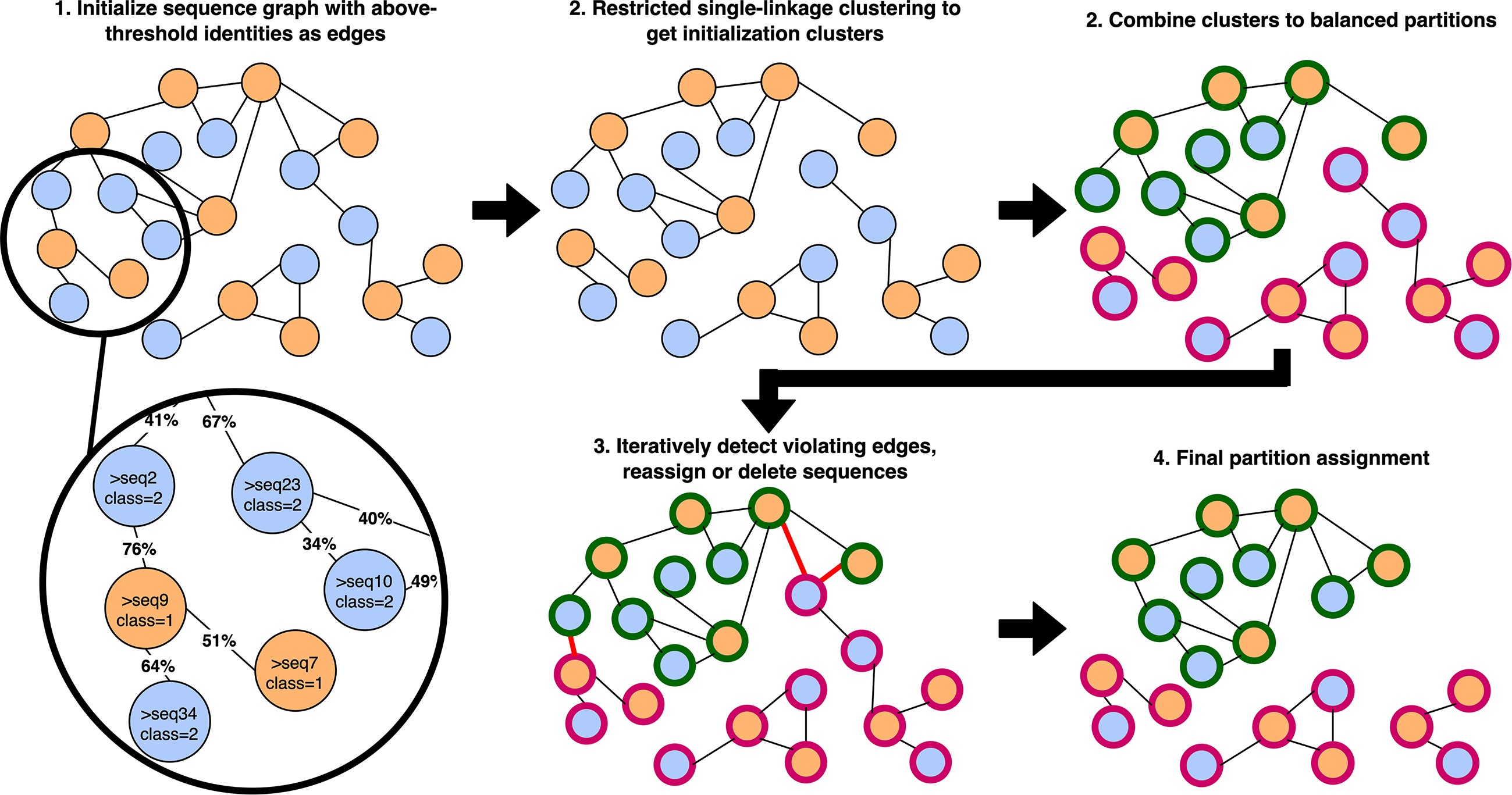 GraphPart: homology partitioning for biological sequence analysis
GraphPart: homology partitioning for biological sequence analysis
NAR Genomics and Bioinformatics, 2023
Felix Teufel, Magnús Halldór Gíslason, José Juan Almagro Armenteros, Alexander Rosenberg Johansen, Ole Winther, Henrik Nielsen
We introduce GraphPart, an algorithm for homology partitioning of biological sequence datasets for machine learning.
Code on Github
Python package
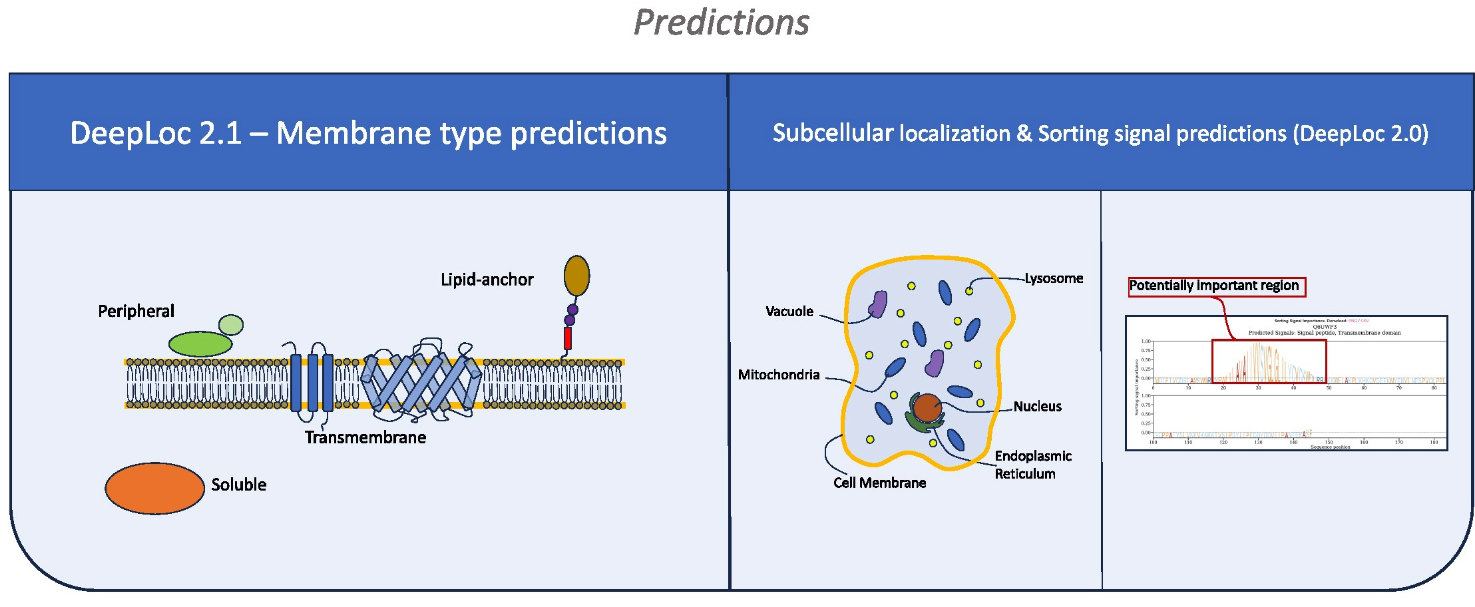 DeepLoc 2.1: multi-label membrane protein type prediction using protein language models
DeepLoc 2.1: multi-label membrane protein type prediction using protein language models
Nucleic Acids Research, 2024
Marius Thrane Ødum, Felix Teufel, Vineet Thumuluri, José Juan Almagro Armenteros, Alexander Rosenberg Johansen, Ole Winther, Henrik Nielsen
DeepLoc 2.1 predicts protein subcellular locations and membrane protein types in eukaryotes.
Available online at DTU Health Tech
Bioactive peptide discovery
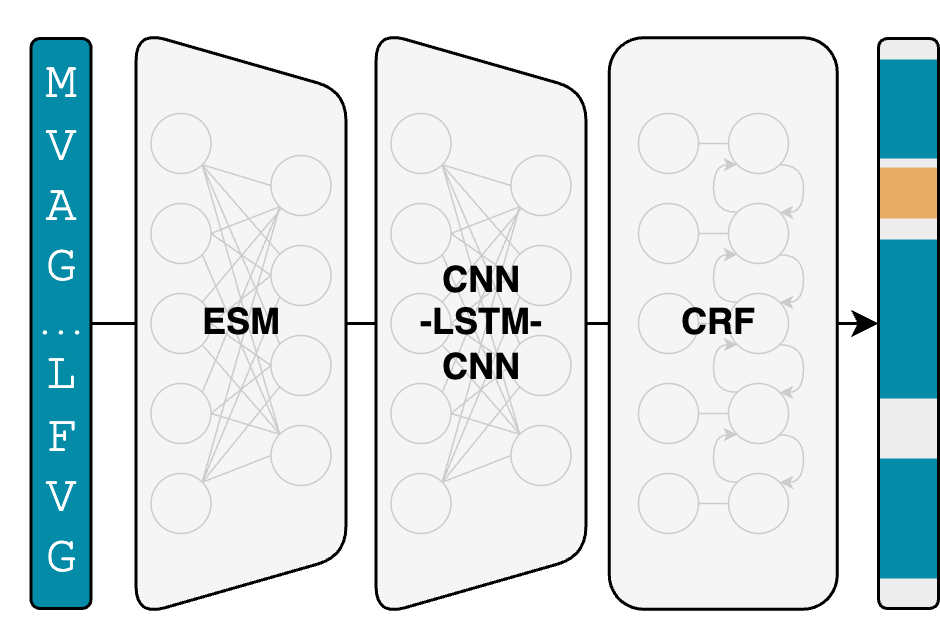 DeepPeptide predicts cleaved peptides in proteins using conditional random fields
DeepPeptide predicts cleaved peptides in proteins using conditional random fields
Bioinformatics, 2023
Felix Teufel, Jan C. Refsgaard, Christian T. Madsen, Carsten Stahlhut, Mads Grønborg, Ole Winther, Dennis Madsen
Predicting native cleaved peptide products directly from precursor protein sequences, without requiring experimental peptide data.
Available online at University of Copenhagen Biolib
Code on Github
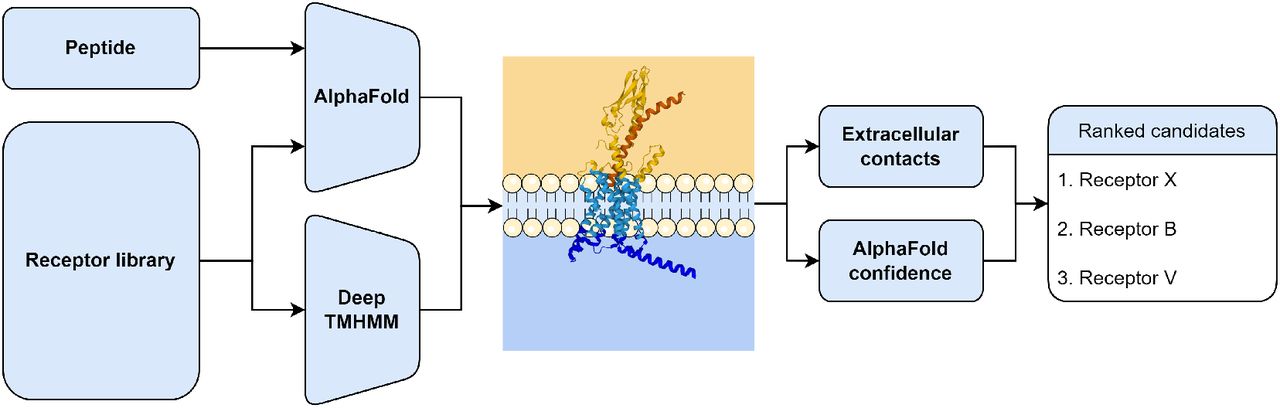 Deorphanizing Peptides using Structure Prediction
Deorphanizing Peptides using Structure Prediction
Journal of Chemical Information and Modeling, 2023 and MLSB Workshop at NeurIPS, 2022
Felix Teufel, Jan C. Refsgaard, Marina A. Kasimova, Kristine Deibler, Christian T. Madsen, Carsten Stahlhut, Mads Grønborg, Ole Winther, Dennis Madsen
We combined AlphaFold-Multimer with DeepTMHMM as a peptide receptor deorphanization method.
Code on Github
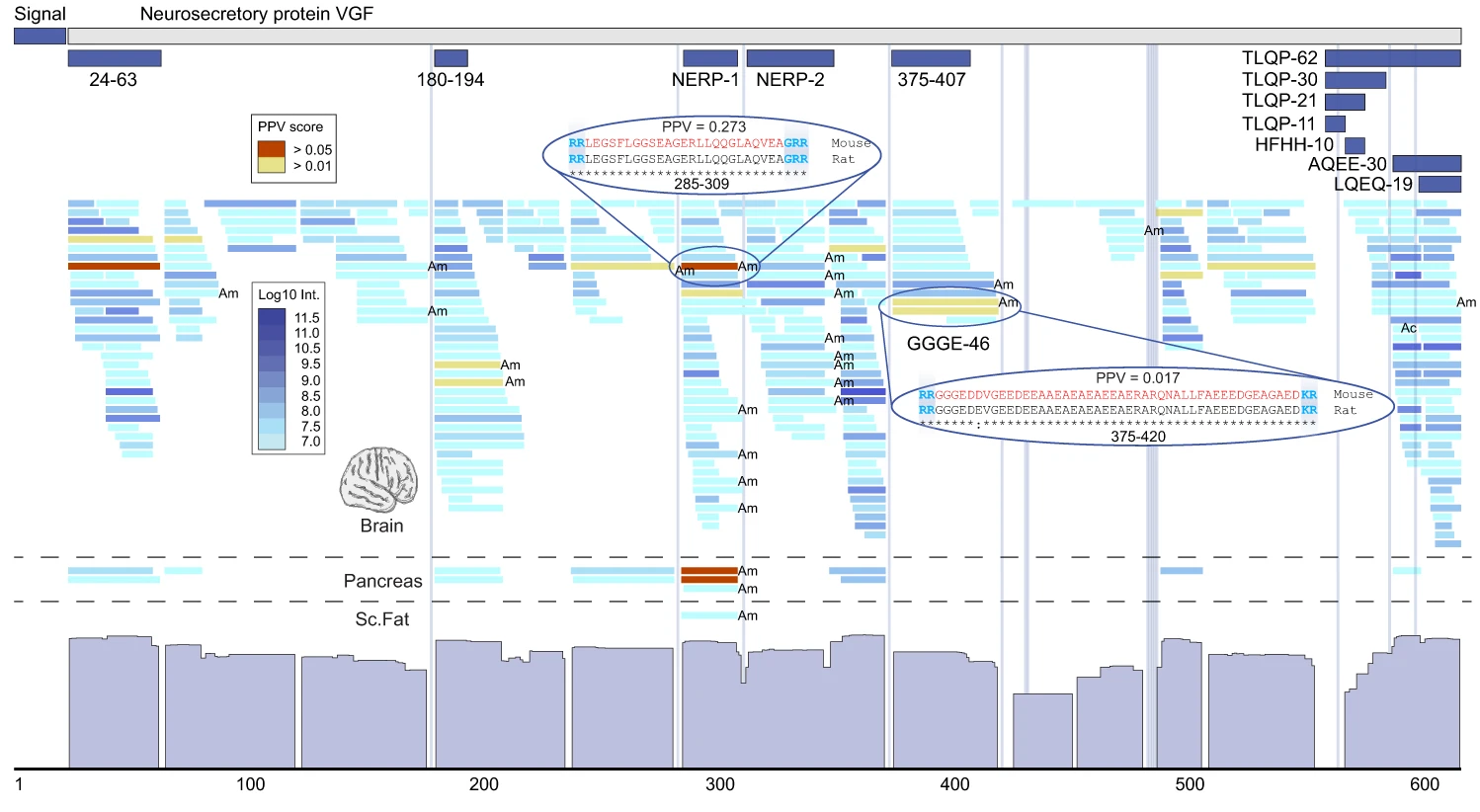 Combining mass spectrometry and machine learning to discover bioactive peptides
Combining mass spectrometry and machine learning to discover bioactive peptides
Nature Communications, 2022
Christian T. Madsen, Jan C. Refsgaard, Felix Teufel, Sonny K. Kjærulff, Zhe Wang, Guangjun Meng, Carsten Jessen, Petteri Heljo, Qunfeng Jiang, Xin Zhao, Bo Wu, Xueping Zhou, Yang Tang, Jacob F. Jeppesen, Christian D. Kelstrup, Stephen T. Buckley, Søren Tullin, Jan Nygaard-Jensen, Xiaoli Chen, Fang Zhang, Jesper V. Olsen, Dan Han, Mads Grønborg & Ulrik de Lichtenberg
Discovering bioactive peptides in large-scale peptidomics data from their mass spectrometry signal.
Code on Github
